(alignment.append() and alignment.check()).
- Generate molecular topology for the target sequence
(model.generate_topology()).
Disulfides in the target are assigned here from the equivalent
disulfides in the templates (model.patch_ss_templates()). Any user defined
patches are also done here (as defined in the
automodel.special_patches() routine).
- Calculate coordinates for atoms that have equivalent atoms in the
templates as an average over all templates (model.transfer_xyz())
(alternatively, read the initial coordinates from a file).
- Build the remaining unknown coordinates using internal coordinates
from the CHARMM topology library (model.build()).
- Write the initial model to a file with extension .ini (model.write()).
- Generate stereochemical, homology-derived, and special restraints
(Restraints.make())
(alternatively, skip this and assume the restraints file already exists):
stereochemical restraint_type = 'bond angle dihedral improper' mainchain dihedrals 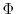 ,
, 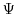
restraint_type = 'phi-psi_binormal' mainchain dihedral 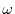
restraint_type = 'omega_dihedral' sidechain dihedral 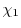
restraint_type = 'chi1_dihedral' sidechain dihedral 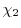
restraint_type = 'chi2_dihedral' sidechain dihedral 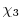
restraint_type = 'chi3_dihedral' sidechain dihedral 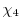
restraint_type = 'chi4_dihedral' mainchain CA-CA distance restraint_type = 'distance' mainchain N-O distance restraint_type = 'distance' sidechain-mainchain distance restraint_type = 'distance' sidechain-sidechain distance restraint_type = 'distance' ligand distance restraints automodel.nonstd_restraints() routine user defined automodel.special_restraints() routine non-bonded pairs distance restraint_type = 'sphere'; calculated on the fly - Write all restraints to a file with extension .rsr (Restraints.write()).
- Generate the optimization schedule for the variable target function
method (VTFM).
- Read the initial model (usually from the .ini file from 2.d)
(model.read()).
- Randomize the initial structure by adding a random number between
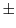 automodel.deviation angstroms to all atomic positions
(selection.randomize_xyz()).
automodel.deviation angstroms to all atomic positions
(selection.randomize_xyz()).
- Optimize the model:
- Partially optimize the model by VTFM; Repeat the following steps
as many times as specified by the optimization schedule:
- Select only the restraints that operate on the atoms that are close enough in sequence, as specified by the current step of VTFM (Restraints.pick()).
- Optimize the model by conjugate gradients, using only currently selected restraints (conjugate_gradients()).
- Refine the model by simulated annealing with molecular dynamics,
if so selected:
- Do a short conjugate gradients optimization (conjugate_gradients()).
- Increase temperature in several steps and do molecular dynamics optimization at each temperature (molecular_dynamics()).
- Decrease temperature in several steps and do molecular dynamics optimization at each temperature (molecular_dynamics()).
- Do a short conjugate gradients optimization (conjugate_gradients()).
- Partially optimize the model by VTFM; Repeat the following steps
as many times as specified by the optimization schedule:
- Calculate the remaining restraint violations and write them out
(selection.energy()).
- Write out the final model to a file with extension .B9999????.pdb
where ???? indicates the model number (model.write()). Also write
out the violations profile.
- Superpose the models and the templates, if so selected by
automodel.final_malign3d = True, and write
them out (alignment.append_model(), alignment.malign3d()).
- Do loop modeling if so selected using the loopmodel class.