



Next: The selection class: handling
Up: The secondary_structure module: secondary
Previous: strand() make
Contents
Index
sheet(atom1, atom2, sheet_h_bonds)
This calculates H-bonding restraints for a pair of
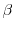 -strands. atom1 and atom2 specify the first H-bond in the
-strands. atom1 and atom2 specify the first H-bond in the 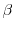 -sheet
ladder. sheet_h_bonds specifies the number of H-bonds to be added --
positive for a parallel sheet, and negative for an anti-parallel sheet.
In a parallel sheet, hydrogen
bonds start at the first or the second term of the following series
(depending on atom1 and atom2): 1N:1O, 1O:3N, 3N:3O, 3O:5N, etc.
For an anti-parallel sheet, the corresponding series is 1N:3O, 1O:3N, 3N:1O,
3O:1N, etc. (note that the residue indices run in decreasing order for the
second strand in this case). The extended structure of the individual strands
themselves is not enforced; use separate strand() restraints if so desired.
-sheet
ladder. sheet_h_bonds specifies the number of H-bonds to be added --
positive for a parallel sheet, and negative for an anti-parallel sheet.
In a parallel sheet, hydrogen
bonds start at the first or the second term of the following series
(depending on atom1 and atom2): 1N:1O, 1O:3N, 3N:3O, 3O:5N, etc.
For an anti-parallel sheet, the corresponding series is 1N:3O, 1O:3N, 3N:1O,
3O:1N, etc. (note that the residue indices run in decreasing order for the
second strand in this case). The extended structure of the individual strands
themselves is not enforced; use separate strand() restraints if so desired.
All of the restraints have the physical.h_bond physical restraint type,
so can be strengthened or weakened by creating a physical.values() object
(see also Section 2.2.2).
To actually add the restraints, pass the new object to Restraints.add().
See Section 2.2.10 for an example.




Next: The selection class: handling
Up: The secondary_structure module: secondary
Previous: strand() make
Contents
Index
Automatic builds
2009-01-30
-strands. atom1 and atom2 specify the first H-bond in the
-sheet ladder. sheet_h_bonds specifies the number of H-bonds to be added -- positive for a parallel sheet, and negative for an anti-parallel sheet. In a parallel sheet, hydrogen bonds start at the first or the second term of the following series (depending on atom1 and atom2): 1N:1O, 1O:3N, 3N:3O, 3O:5N, etc. For an anti-parallel sheet, the corresponding series is 1N:3O, 1O:3N, 3N:1O, 3O:1N, etc. (note that the residue indices run in decreasing order for the second strand in this case). The extended structure of the individual strands themselves is not enforced; use separate strand() restraints if so desired.